Hi everyone,
I want to create a dataframe from another one using python code. My dataframe that I want to modify look like that :
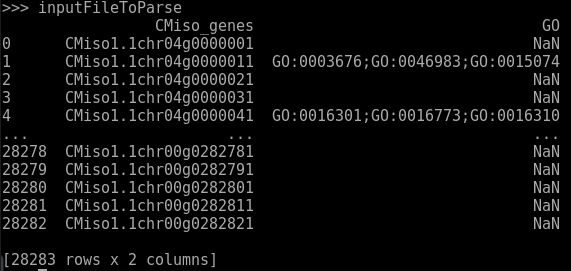
I want to produce from this dataframe : (I take just the first 3 rows of the dataframe to build the exemple)
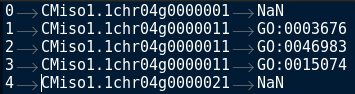
As you can see I want to separate the Go term per gene in order to build separate goterm list for CC,MF and BP.
this is the function that I start to build but I am block because of the mix of float and string value that not allow to separate the GO column.
sep1="\t"
sep2=","
sepIn=";"
# Read input files
inputFileToParse=pd.read_csv(inputFileToParsePath, sep=sep1)
pivotFile=pd.read_csv(pivotFileForParsingPath,sep=sep2, index_col=0)
# Erasing unwanted column
del pivotFile["level_0"]
# get columns from the input dataframe
inputFileColumnsNames=inputFileToParse.columns
# function
def getListDescGoPerProcess(inputFileToParse,pivotFile,sepIn)
# separate all row if multiple value
if inputFileToParse['GO'].str.contains(';'):
print("some gene as multiple GO term, it will be separate for the parsing")
pd.concat([pd.Series(row['CMiso_genes'], row['GO'].split(';'))
for _, row in inputFileToParse.iterrows()]).reset_index()
# Create the 3 output dateframe to save
BPtable=pd.DataFrame(columns = inputFileColumnsNames)
CCtable=pd.DataFrame(columns = inputFileColumnsNames)
MFtable=pd.DataFrame(columns = inputFileColumnsNames)
if
I have not finish the function already because I am block on the lign when I concatenate the sliting of the second colums. But as I said before the function split cannot take in count float value.
Do you have any suggestion to build this dataframe ?
Thanks in advance


Can you please post the input data as text using the
codeenvironment. It's too complicated for people to create by hand from an image.Hi, thanks for your respons.
Here you have the dataframe as text :