Hi, I have a question for ChIP-seq peak calling. Because macs2 fits a bimodal peaks to find the combination site. Thus, is it true that I need to see a bimodal peak in IGV? But actually I just saw a unimodal peak in IGV, so that's why? Thanks a lot!
What macs does is to pile up the 5' ends of the reads and then extend them to 3' by the estimated average fragment size. Since the 5' ends accumulate on either side of the actual protein binding location it then shifts these pileups to 3' end to infer the actual binding site, see also https://hbctraining.github.io/Intro-to-ChIPseq/lessons/05_peak_calling_macs.html and read the original macs paper which describes this in much detail.
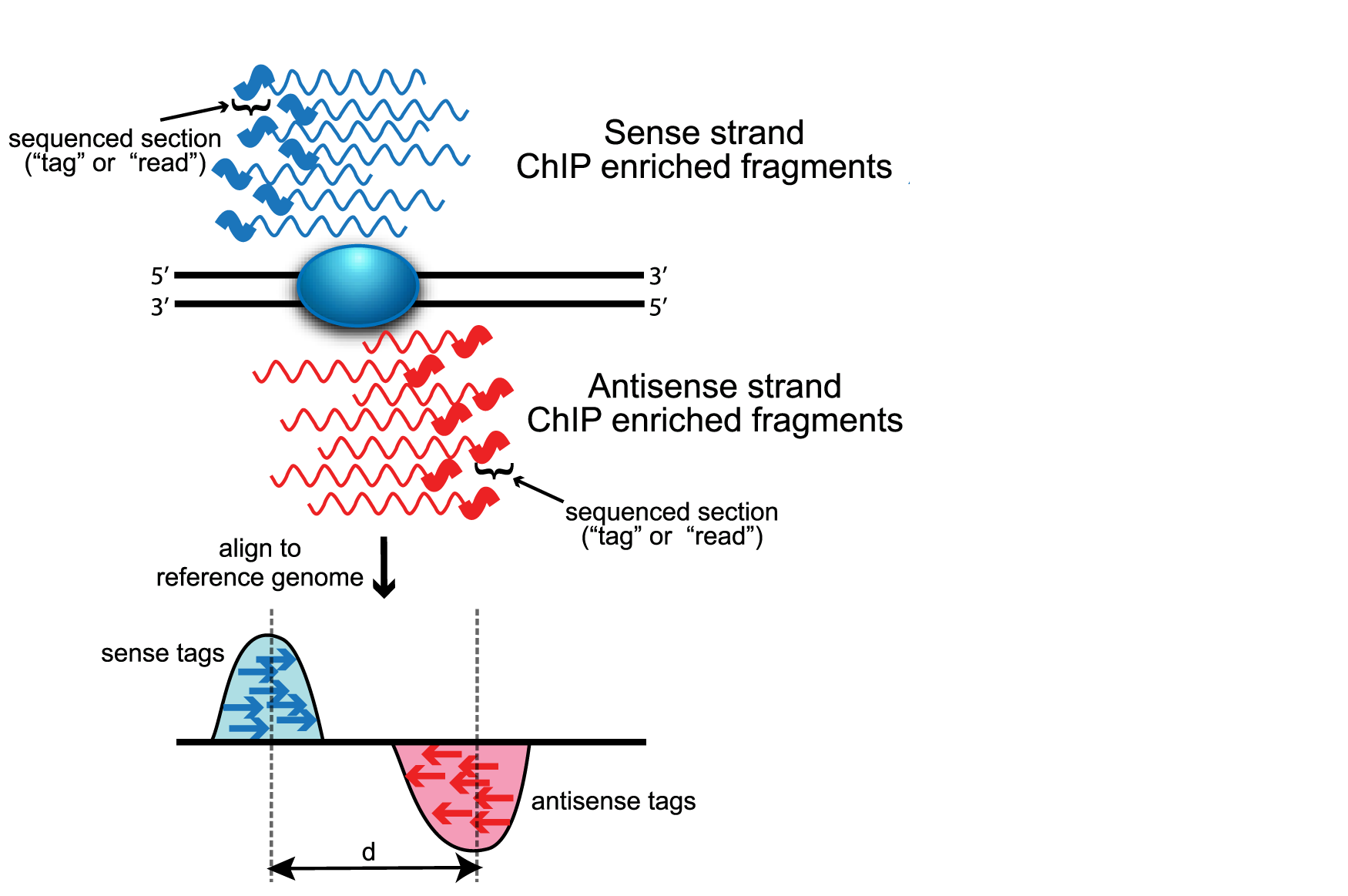
You don't see a bimodal peak because in a browser track the reads are usually extended to fragment size and not reduced to the 5' ends. If you made a track of only like a few bases starting from the 5' end of reads you might see it, never tried that actually.
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.

