Hi, I decided to use Trimmomatic for trimming raw reads. I saw many researchers used the inputs (LEADING:3 TRAILING:3) authors suggested on their webpage (https://www.usadellab.org/cms/?page=trimmomatic) for trimming RNA-Seq reads.
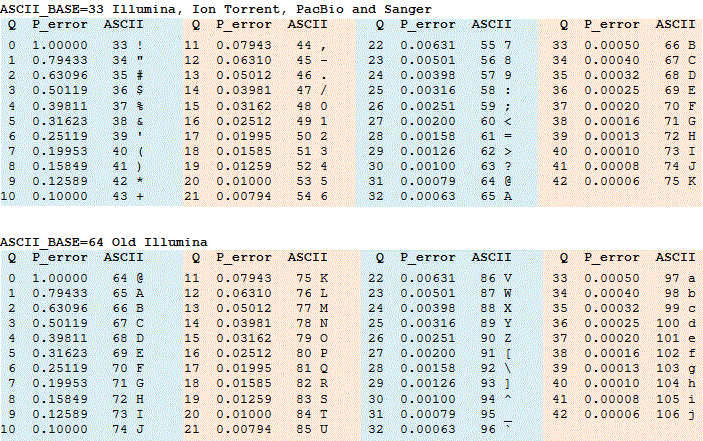
I thought these values are very low, so I started to find out their exact meaning. A website (http://drive5.com/usearch/manual/quality_score.html) used the below figure to explain the Phred (quality score), and stated that: "Note that a Q score of 3 means P (the error probability)=0.5, meaning that there is a 50% chance the base is wrong, and lower values represent even higher probabilities of error." Can anyone explain why you suggested such low criteria for filtering reads? Best, Armin


First of all do you have a specific problem with beginning or ends of reads as far as quality? If you don't then you don't have to strictly trim them. Poor quality bases (if they are adapter/contaminants) can be soft-clipped by aligners so don't strictly require trimming. That said, if you are planning to do any de novo assembly work then trimming the data more stringently at Q15 or higher may be warranted.
Hi, Thank you for your reply. That is another problem I still have. Some scientists suggested removing the first few bases according to the "Per base sequence quality" and "Per base sequence content" sections of FastQC, while others suggested leaving these bases. I uploaded the results for one of my reads ( and
and  ). I want to use Kallisto, which does a pseudo alignment to the reference transcriptome. I decided to trim the first 12 bases.
However, in this post, I asked another question. I want to know why the authors of Trimmomatic suggested such low criteria on their website. Unfortunately, I was not able to contact the authors.
). I want to use Kallisto, which does a pseudo alignment to the reference transcriptome. I decided to trim the first 12 bases.
However, in this post, I asked another question. I want to know why the authors of Trimmomatic suggested such low criteria on their website. Unfortunately, I was not able to contact the authors.
Please read these blog posts from authors of FastQC which should address your questions.
https://sequencing.qcfail.com/articles/libraries-can-contain-technical-duplication/
https://sequencing.qcfail.com/articles/positional-sequence-bias-in-random-primed-libraries/