Hi everyone,
I have a question on basic statistics. I feel really bad about asking this but I'm having a brain freeze now. So, I'm going back to basics.
Purpose: Calculate expected trinucleotide frequencies specific to 5'UTR regions only, which will be used to check whether some windows (around 20bp long) at particular locations (such as beginning or end) in a set of specific 5'UTRs have different amount of these trinucleotides by calculating observed-to-expected ratio.
Calculated Data: I collected all 5'UTR sequences and counted all nucleotides. I have calculated their frequency as follows:

where  ,
, 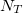 ,
, 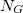 ,
, 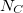 represents occurrence count of each nucleotide in all 5'UTR regions (these regions only!) and
represents occurrence count of each nucleotide in all 5'UTR regions (these regions only!) and 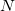 represents total amount of nucleotides in all 5'UTR regions.
represents total amount of nucleotides in all 5'UTR regions.
From this I calculate expected frequency of a trinucleotide by
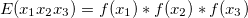
where 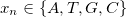 . Then, for each window of interest, I count the occurrence of each
. Then, for each window of interest, I count the occurrence of each 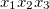 trinucleotide and calculate observed frequency by
trinucleotide and calculate observed frequency by
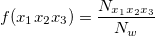
where 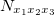 represents the occurrence of
represents the occurrence of 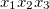 trinucleotide and
trinucleotide and 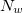 represents the window size.
represents the window size.
Lastly, observed-to-expected ratio becomes
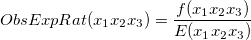
So, I feel like I'm thinking something extremely wrong in this basic thing and I should take window size in the expected frequency calculation as well. But I couldn't be able to convince myself to do this since I already get a "frequency" on the observed part as well.
Am I doing something really wrong and misinterpreting/overthinking everything now?
EDIT: I uploaded a couple of images to a more compatible website but decided it wasn't worth the effort - Ram.


Thanks a lot! Yes, I look for strand effect as well by doing same calculation for the sense strand and anti-sense strand separately first, then I'm getting a sum of occurrences in both strands and divide by
window_size*2and calculate a obs-to-exp ratio from that as well.