You can do this with karyoploteR in combination with ggplotify and cowplot.
You'll have to create two independent karyoplots (here contained in two functions) and use ggplotify and cowplot to combine them into a single image.
The first karyoplot is just the chromosome ideogram with the rectangle for the zoomed-in region and the second one is the one with the actual data. In the second one you can plot rectangle representing SNPs, but you can also plot any other data or annotations using other karyoploteR functions
In any case, here's the code
library(karyoploteR)
library(ggplotify)
library(cowplot)
snps <- toGRanges(c("chr7:21e6-21e6", "chr7:21.2e6-21.2e6", "chr7:21.4e6-21.4e6"))
snps$name <- c("rs0002", "rs0004", "rs0006")
snps$y0 = c(0, 0.5, 0)
snps$y1=c(0.3, 0.8, 0.3)
zoom.reg <- toGRanges("chr7:20.5e6-23e6")
plotChromosome <- function() {
pp <- getDefaultPlotParams(plot.type = 6)
pp$topmargin <- 20
pp$bottommargin <- 20
kp <- plotKaryotype(chromosomes = "chr7", plot.type=6, plot.params = pp, cex=1.4)
kpAddBaseNumbers(kp, add.units = TRUE, cex=1.8)
kpRect(kp, zoom.reg, y0=-0.4, y1=1.4, col=NA, border="red", lwd=2)
}
plotZoom <- function() {
pp <- getDefaultPlotParams(1)
pp$bottommargin <- 20
pp$topmargin <- 20
kp <- plotKaryotype(zoom=zoom.reg, plot.params = pp, cex=1.8)
kpAddBaseNumbers(kp, tick.dist = 1e6, minor.tick.dist = 0.2e6, add.units = TRUE, cex=1.4)
kpRect(kp, snps, col="steelblue", border="steelblue")
kpPlotNames(kp, snps, labels = snps$name, pos="left", cex=1.6)
kpAddLabels(kp, labels = "Selected SNPs", srt=90, pos=3, cex=1.8)
}
png("General_plus_zoom.png", width=1000, height=500)
p1 <- as.ggplot(expression(plotChromosome()))
p2 <- as.ggplot(expression(plotZoom()))
plot_grid(p1, p2, ncol=1, rel_heights = c(1,3))
dev.off()
And this will give you an image like this
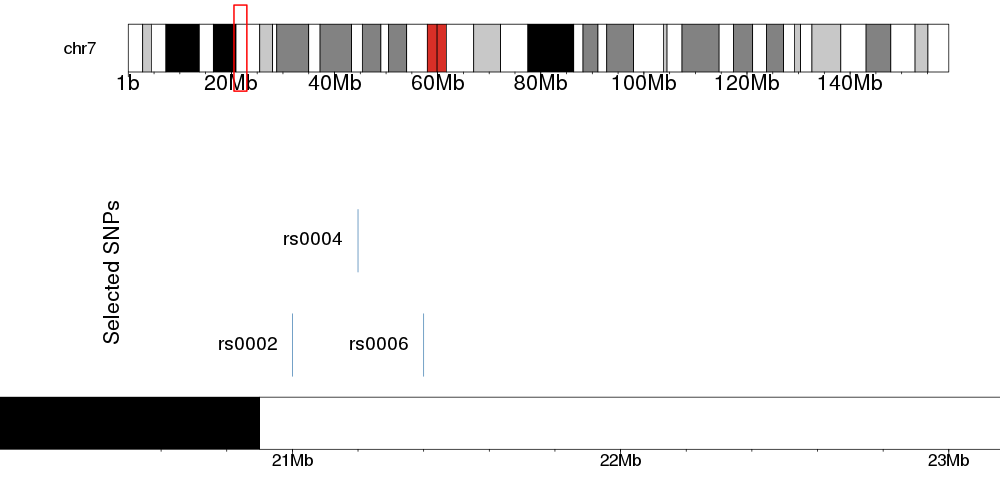
You can improve, modify and customize the image however you want. You can find more information in the karyoploteR tutorial.
Hope this helps




Hey, did you figure this out? I'm trying to do the same. I found a package called "mapsnp". But it is not available in the current version of R (3.6.1) :(