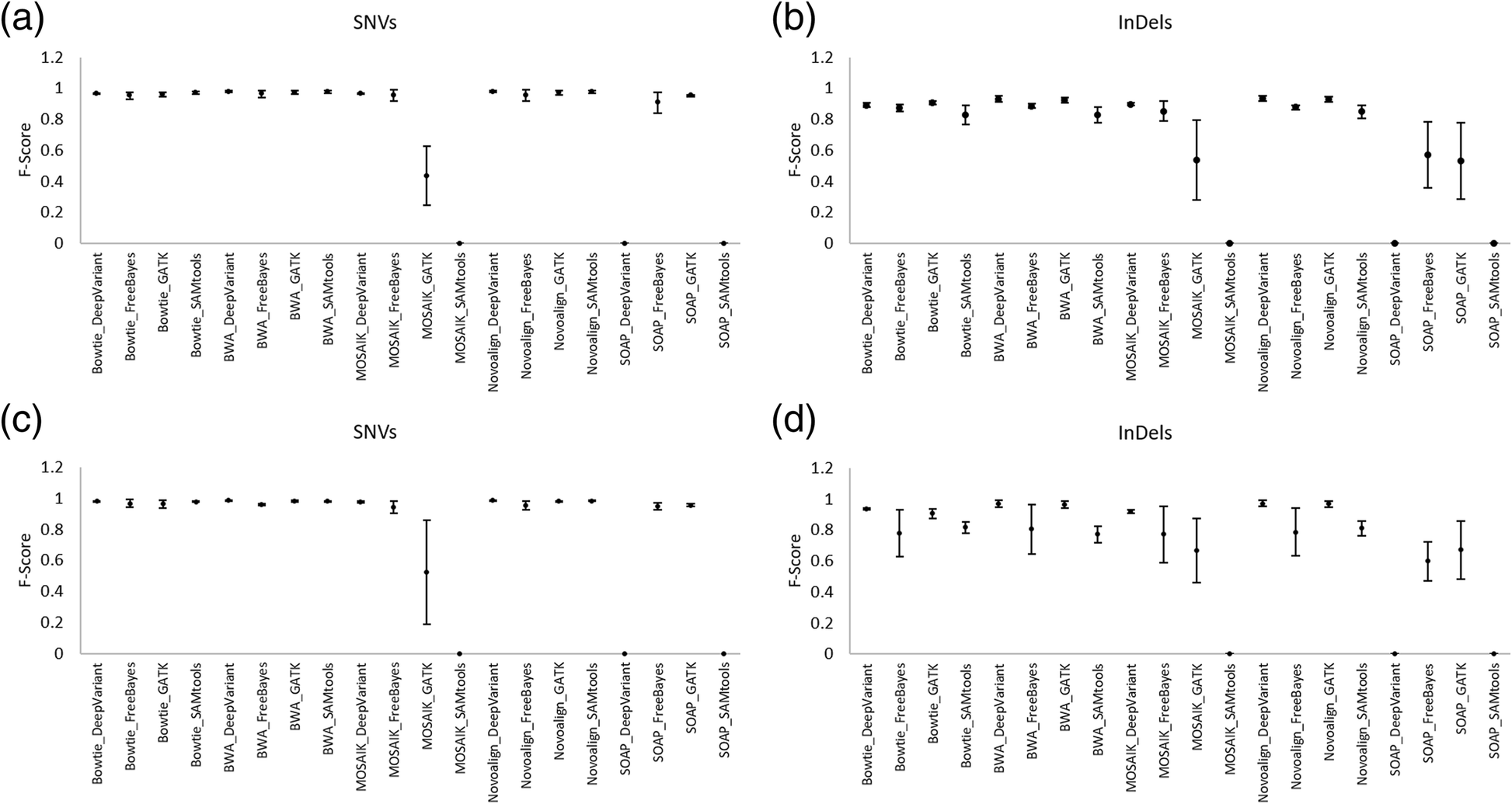
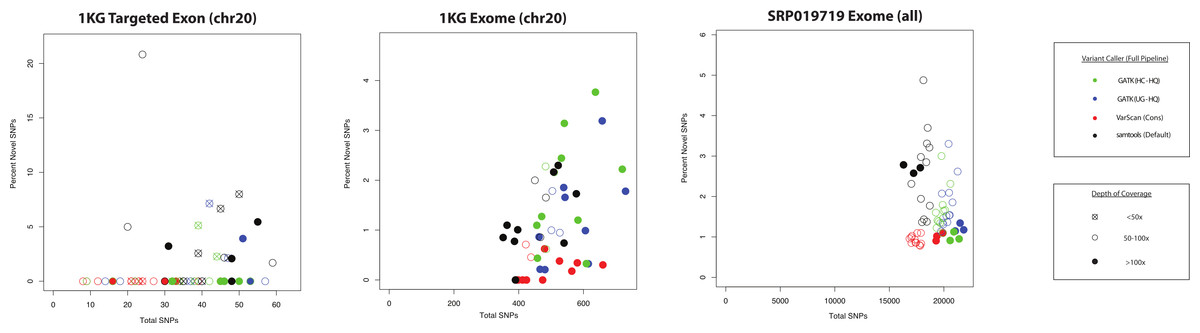
Dear *,
I currently try out DeepVariant, which was designed for Whole Exome Sequencing and Whole Genome Sequencing.
Now I wonder if there is any problem, if I would use it for gene panels, so a subset of whole exome sequencing (more or less).
Best regards, Daniel