Hello guys, Im very familiar with Broad institute portal, but now since i need to work with molecular profiles and clinical drug response data. now i learned how to obtain clinial drug response data from GDC portal https://portal.gdc.cancer.gov/ my cancer of interest is colorectal cancer (COADREAD) , but i found that this type of cancer is not availabe in this GDC portal ( primary site). i only find colon (coad) instead, is this right?
Another issue, if i want to get clinical drug response data from GDC portal and the molecular profiles of the same patient id from Broad insitutue firhose is this biologically reasonable and have no effect on the results or the quality control?
Appreciate any help!

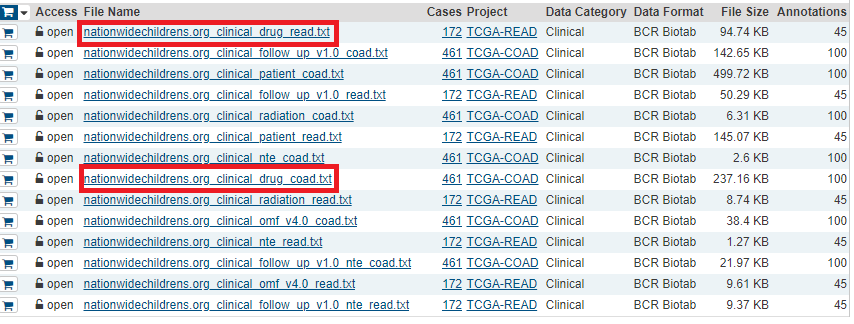
 Gene expression:
Gene expression:

please, Any help or any hints or a tutorial to read about this issue if that possible?