Entering edit mode
5.0 years ago
arshiaurora
▴
10
Hello,
I would like to introduce a light visualization R package to plot and clustered `omics data like categorical (mutations. other clinical variables), or continuous (gene expression, age, etc) variables: panelmap
- panelmap creates appropriate legends for all the variables used and provided complete flexibility and various customizations of the appearance of the plot.
- makepanel A one-line command that takes in the group variable of interest along with other variables to see their distribution among each group. makepanel also allows for appropriate statistical tests to check for associations and returns a summary table of all the variables used.
See an example on mtcars dataset-

- circomap circomap allows us to visualize more than one datatypes in a circular layout consisting of multiple panelmaps. For instance, if I repeat data used above 3 times, then I can visualize this via circomap by arranging 3 panelmaps in a circular fashion.
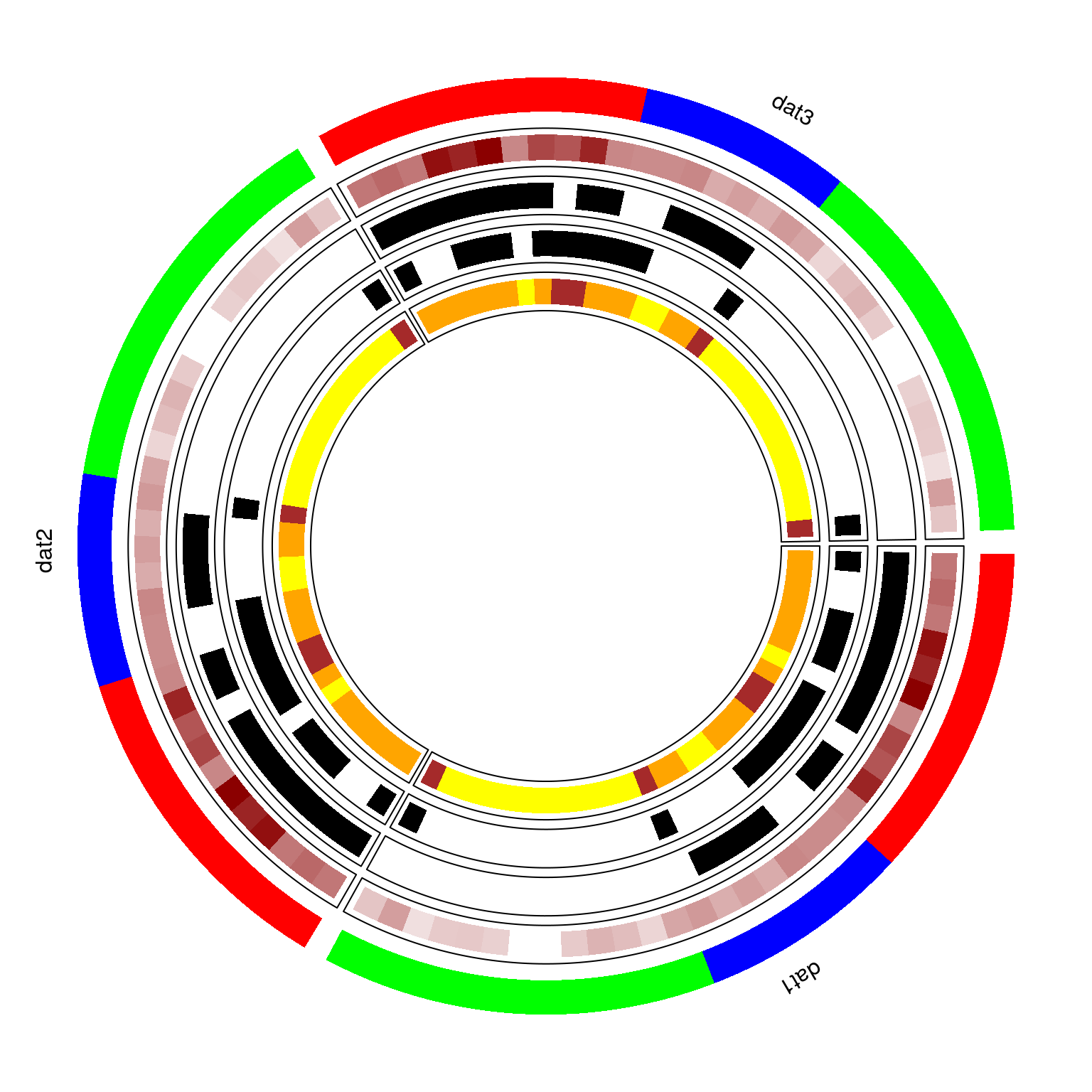
- makepanel_order This function plots a panelmap when no group is provided. Only works for binary or mutation data for now. One can also provide a
userdefined order to plot other types of variables lacking a group.
panelmap was used to create figures for this manuscript
To learn more about panelmap see this and a detailed tutorial can be found here

