Entering edit mode
3.8 years ago
keremwai
▴
10
Hi,
What would you advise the best cloud environment for genetics? I would love to just hear what is it, and if you feel like please add why. My needs for the time are mainly for RNA-seq, alignment and so on.
Thank you in advance,
Kerem


The term "cloud" can mean a lot of things, but a loose definition would mean that the programs and databases are on a server, but you run them from the desktop. You get the full power of the server, with the convenience of working from any computer you happen to be on. HOW you access the server can make a difference.
Many sites use the Galaxy https://usegalaxy.org/, which lets you run tasks and workflows on the server, through your web browser. You can try out Galaxy free at this site, but for any substantial work, you need to install Galaxy on a local server at your institution. In my opinion, web-based tools are awkward to use, compared to systems in which you have a full point and click desktop session running on the server. The interaction is slower, and the web browser interface is designed to take up the entire screen, which tremendously impairs your sense of working with the data. To me its more like asking someone else to do each step, and having to explain to them how to do that step, rather than doing it yourself with a few clicks.
Amazon has a commercial genomics environment as part of their broader AWS environment https://aws.amazon.com/health/?nc=sn&loc=1. I haven't used this myself, but their videos show that you can run a complete graphical desktop on their cloud, with the desktop appearing on the screen of any device, whether it be PC, laptop, or even a phone, if you can dock it so that you have a keyboard, monitor and mouse. This is really only useful for genomics if you have a substantial budget. The entry level desktop is no more powerful than a PC you'd buy in a store (eg. 1 or 2 virtual CPUs, maybe 16 Gb RAM etc.) To get an environment useful for Omics work such as RNAseq, you need to spend extra, although the benefit is you can increase or scale back your resources as you need them.
You weren't too specific about what you want to do, but a large number of the common tasks can be done in the BIRCH system http://home.cc.umanitoba.ca/~psgendb. BIRCH packages together hundreds of the most common bioinformatics applications, unified through the BioLegato family of desktop applications.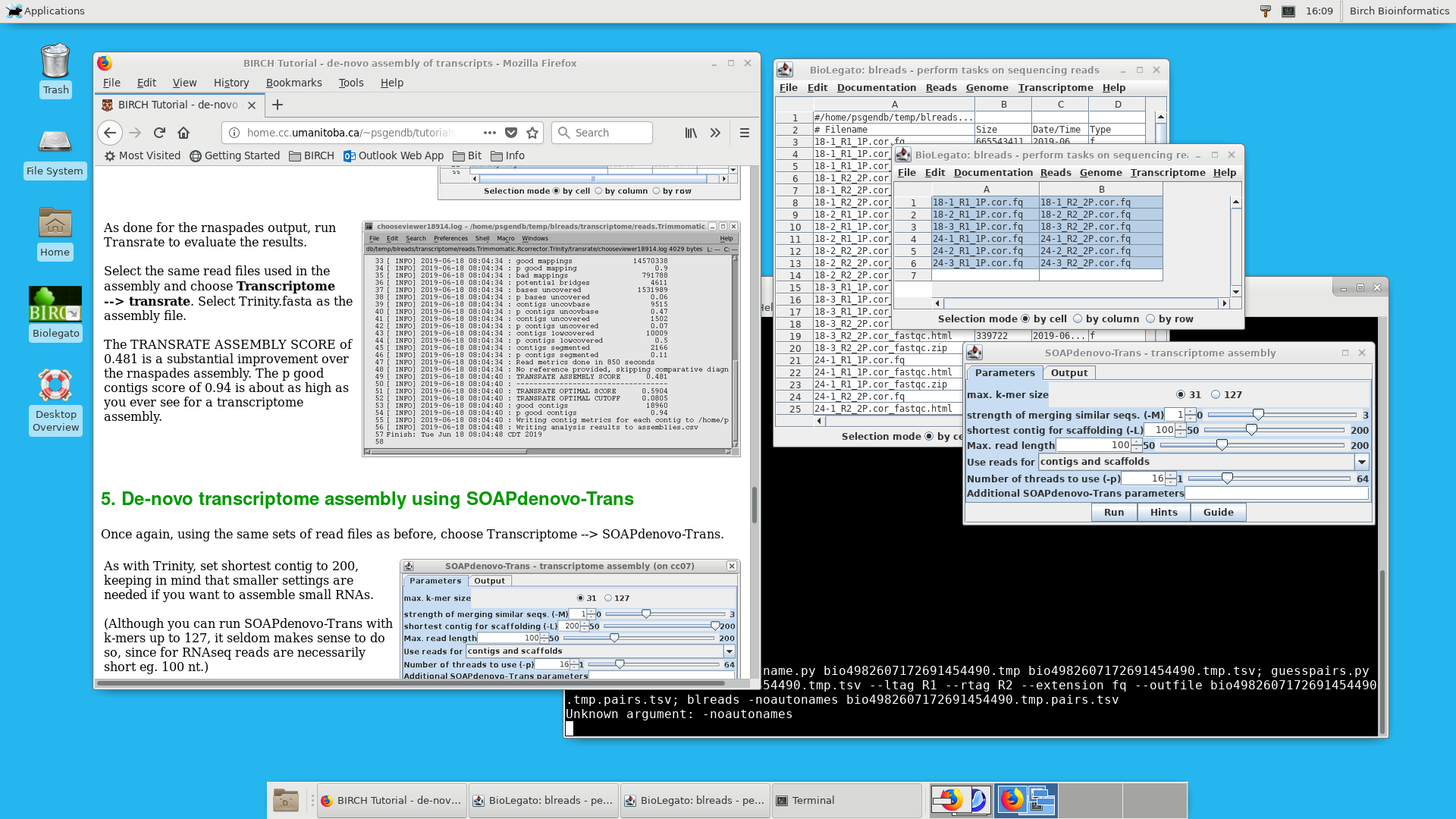 .
.
For smaller tasks, BIRCH can run on a modest desktop computer running Linux or MacOSX. If your institute or department has a more substantial Linux server, you can install BIRCH on the server, and the whole lab group can use BIRCH through a remote desktop client, such as Thinlinc from Cendio https://www.cendio.com. Again, this lets you use a powerful server, from any device.
You can see BIRCH in action on the BIRCH YouTube Channel https://www.youtube.com/channel/UC9_3TfH3sjE0YdToVMChq-w?view_as=public
It depends on the details of what you want to do and how you want to do it and also on other constraints you may have (e.g. how much money you can spend, who is going to do the deployment...). All cloud providers give you more or less the same thing, i.e. access to virtual machines and container orchestration systems but you generally need to take care of the deployment yourself. Setting up a virtual workstation is easy but setting up a cluster is much less straightforward. There's been previous discussion on cloud computing here, check the list of similar posts on the side of this page. Also you may be interested in this review. I'd give a try to public Galaxy instances (e.g. usegalaxy.org for the US or use.galaxy.eu for Europe) and eventually set one up in the cloud if that satisfies your needs.