I need to draw "lollipop", where I could set the length of both vertical lines and the loop size. It shall look similarly to the two right lollipops/RNA secondary structures on this figure:
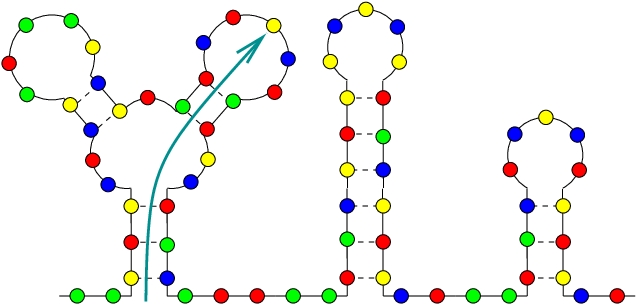
I found a lot of software that can draw beautiful RNA fold figures, given that sequence of nucleotides is provided (eg http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi). On the other hand, I need to just draw this structures - one possibility probably would be to come up with a sequence that produces the exact size of the arms and loop that I want, but I would rather use some R package to do it, if available. Therefore I need script solution as I don't want to draw by hand.
I hope this question is not too off-topic for biostars - if yes, I can certainly move/delete my post:-)


If you do end up having to draw by hand I can recommend a network drawing tool yED that would make the process a lot less painful. Plus you can pregenerate the input data nodes with proper coloring with a small script that generate a graphml input
http://www.yworks.com/en/products_yed_about.html