Does anyone know a good GWAS visualisation package that can be integrated into a website? I've seen a website using this one:
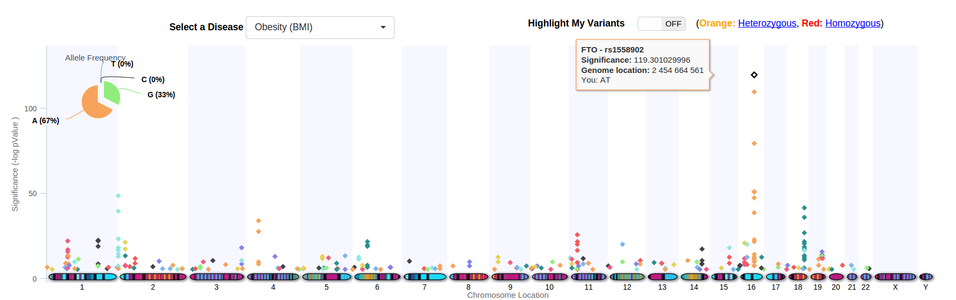
Which looks brilliant. Does anyone know if that is a downloadable package? I'm looking for a package preferably in javascript. I've seen jbrowse but the graphs aren't that nice looking.

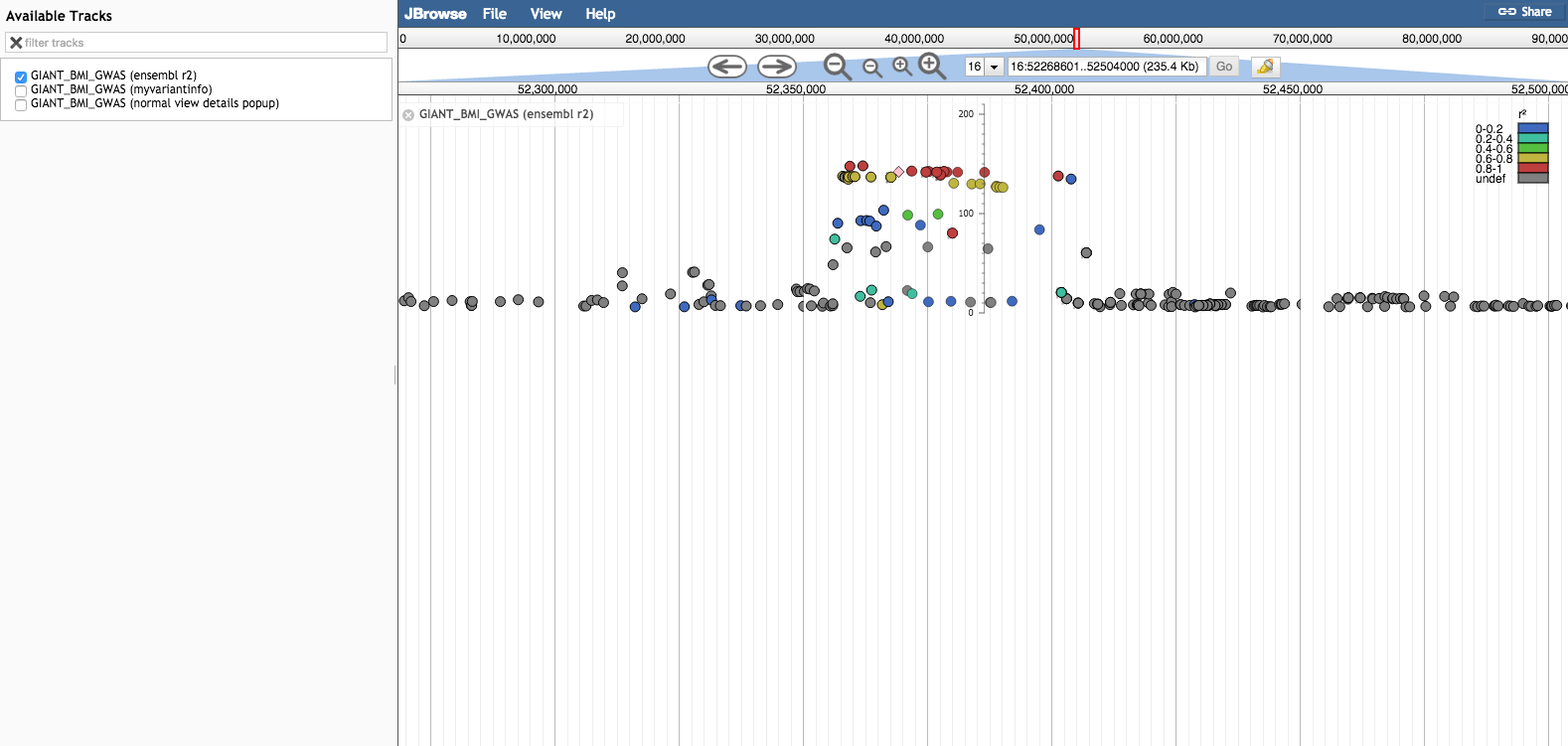

If you don't mind a bit of coding, using R Shiny this should be pretty easy.