Entering edit mode
7.2 years ago
lsp03yjh
▴
860
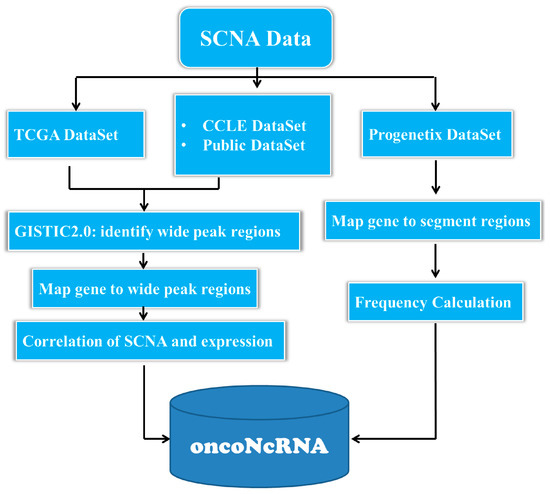
We had released oncoNcRNA, a web portal for explore the ncRNAs (lncRNAs, miRNAs, piRNAs, snoRNAs, tRNAs) with oncogenic potentials in 64 human cancer types(>20000 tumor samples and >800 cancer cell lines). It is a valuable tool for investigating the function and clinical relevance of ncRNAs in human cancers.
oncoNcRNA web site: http://rna.sysu.edu.cn/onconcrna/
- The portal characterizes the somatic copy number alterations (SCNAs) and expression of over 58,000 long non-coding RNAs (lncRNAs), 34,000 piwi-interacting RNAs (piRNAs), 2700 microRNAs (miRNAs), 600 transfer RNAs (tRNAs) and 400 small nucleolar RNAs (snoRNAs) in 64 human cancer types by integrating the data from The Cancer Genome Atlas (TCGA), Cancer Cell Line Encyclopedia (CCLE), Progenetix, and several independent studies.
- We have discovered a large number of ncRNAs which are frequently amplified or deleted within and across tumor types using oncoNcRNA portal.
- We built a web-based tool, Correlations, to explore the relationships between gene expression and copy number from ~10,000 tumor samples in 36 cancer types identified by The Cancer Genome Atlas (TCGA).
- oncoNcRNA provide the expression profiles of lncRNAs, protien-coding genes and other ncRNAs generated from >10000 RNA-seq samples.
oncoNcRNA paper is available at http://www.mdpi.com/2311-553X/3/1/7