Dear all,
I have some libraries with its replicates. To know how similar are they (between replicates) I made a Venn diagram of unique sequences showing this results:
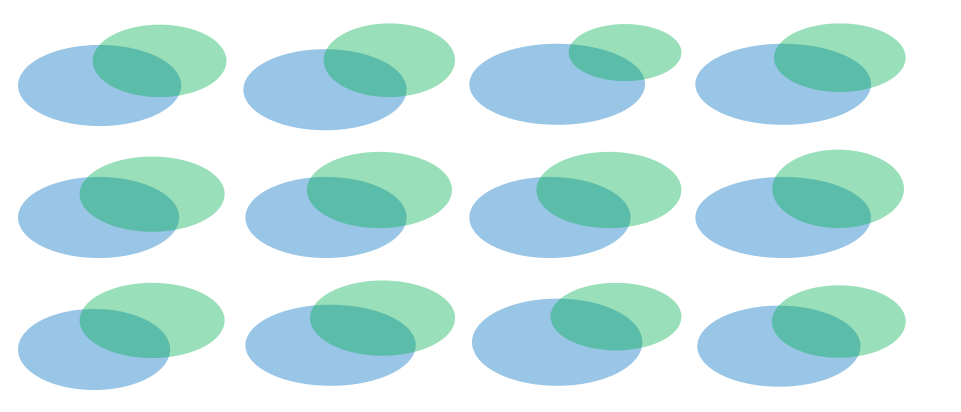
Having that I wondered if removing those sequences with one read would improve the diagram. The result of doing that is the following:
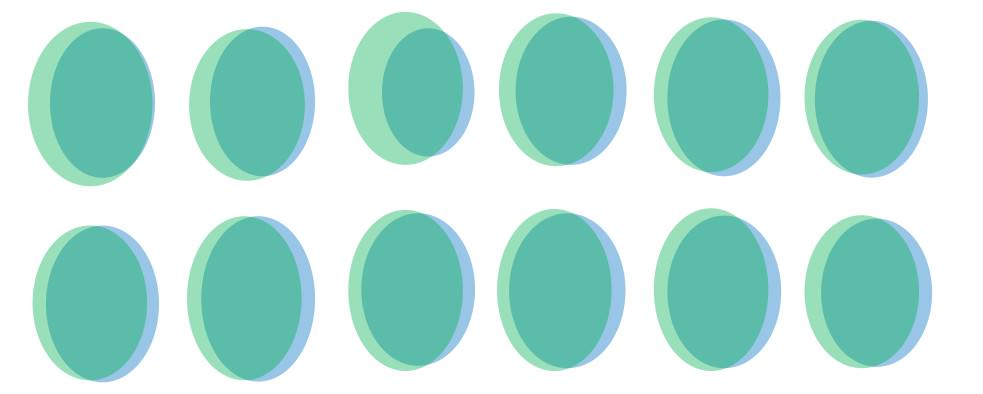
Now I would like to know if there is enough similitude between both replicates. There is a statistical test that I could apply to know this? If not there is some method that I could apply to know if I should remove more sequences with more reads o leave it as it was before?
Thank you everyone
Edit: The libraries come from a smallRNA-seq and I mapped the reads against the genome before doing the Venn diagrams.


Please see How to add images to a Biostars post and follow the guide to add images properly.
What exactly do you want to test, i.e. what is the question of interest here and what do the different pairs represent?