Entering edit mode
6.7 years ago
Chaimaa
▴
260
Hello guys,
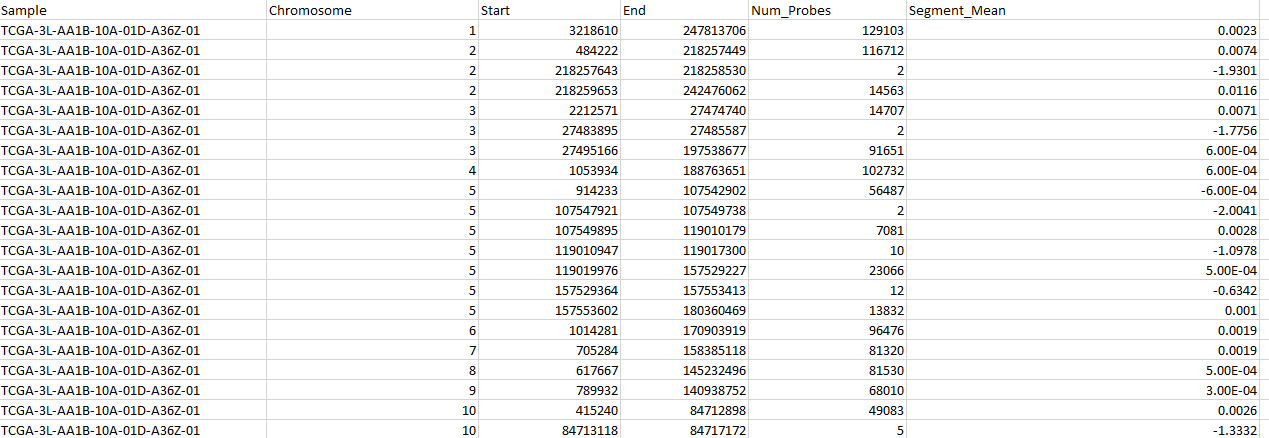
want to convert my cancer data which I got from the Broad Firehose (http://gdac.broadinstitute.org) into a matrix (where the rows will represent the samples and the columns represent the genes).
However, in this data, there are many samples and each sample has many chromosomes(1-23) and many segments mean values
So, which samples have to take among the 23 chromosomes and which segment value?
I appreciate any help!


Please do not add tabular text as a screenshot. You can:
cat tab_sep_file | column -ts $'\t'and copy-paste the output to a code-formatted block here; orcolumncommand as shown above for proper formatting), upload it to gist and add a link to the gist here