When analyzing clinical genomes, I often find it hard to visualize the genomic context of a potential driver mutation (say a point mutation on a key cancer gene). For example, is there simultaneous LOH/amplification at the mutation locus? How many copies of the mutant allele are there? Did the mutation arrive before/after the overlapping copy number change? Is there a structural variation breakpoint near by? Are there other point mutations close by?
Thus, I wonder if there is a plotting tool that visualizes a local range of the genome (e.g. a window big enough to inspect a gene) that integrates (in one plot):
- Point mutations in the region
- The local copy numbers (major/minor allele if available)
- SVs affecting the region
Note that the purpose is more to infer the consequence of a mutation as opposed to validating whether the mutation is real or not, which we can already do with a variety tools such as samtools tview.
Circos is a nice format that integrates all three classes of mutations, but it is more for global scale inspections and does not provide enough resolution to zoom into specific gene regions in order to answer the above questions about key mutations of interest. Another close one is IGV, but it does not incorporate allelic specific copy number info.
Thanks!

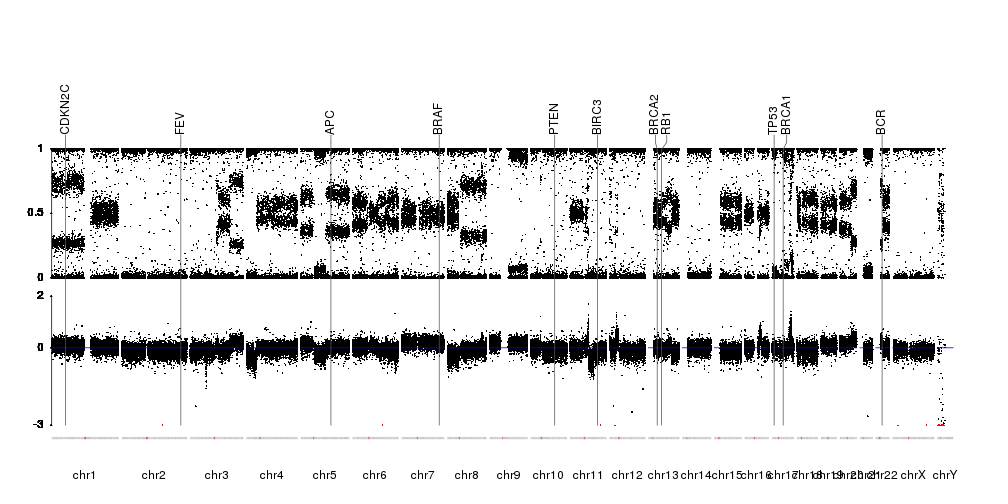
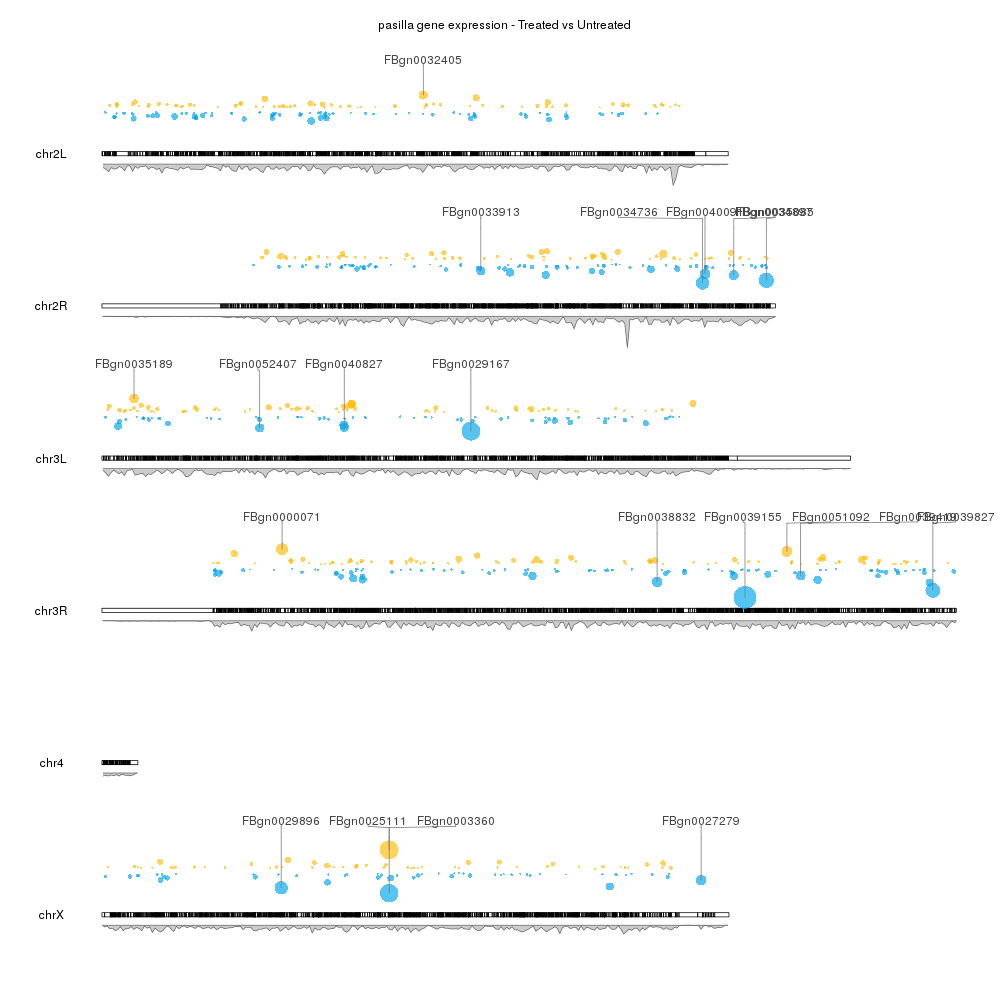
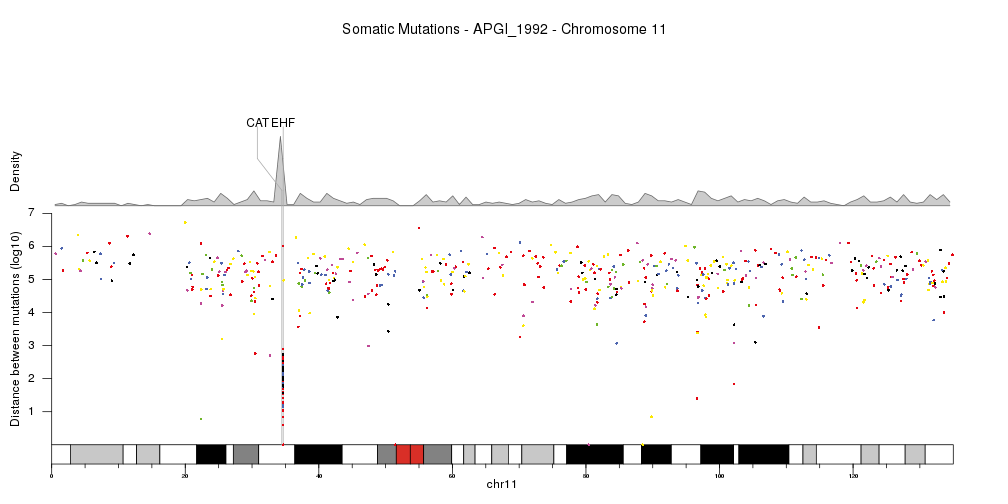

This is a really neat solution!