As you are probably well aware the world has been rocked by the outbreak of a novel coronavirus. Genomic sciences have been at the forefront of identifying and diagnosing the virus; sequence analysis the primary means for tracking its origin and evolution.
You may wonder, what exactly takes place when investigating a novel viral outbreak? What evidence backs up each statement?
To answer all these questions we have embarked on writing a new volume in the Biostar Handbook, a volume titled:
In this new book, we prepare readers to take on the challenges of investigating a novel viral outbreak. By utilizing the latest data and most up to date techniques we will demonstrate procedures, evaluate then validate various statements made in media, then explore other characteristics of the data. Among the subjects that we cover:
- Does the virus have a single origin?
- Has the viral sequence evolved since the outbreak?
- How can we identify the "initial" virus?
- Did the virus jump from other organisms?
- What does the data consist of? Where can we obtain it?
The book is a well documented and comprehensive take on a subject that already had and will continue to have immense impact on society.
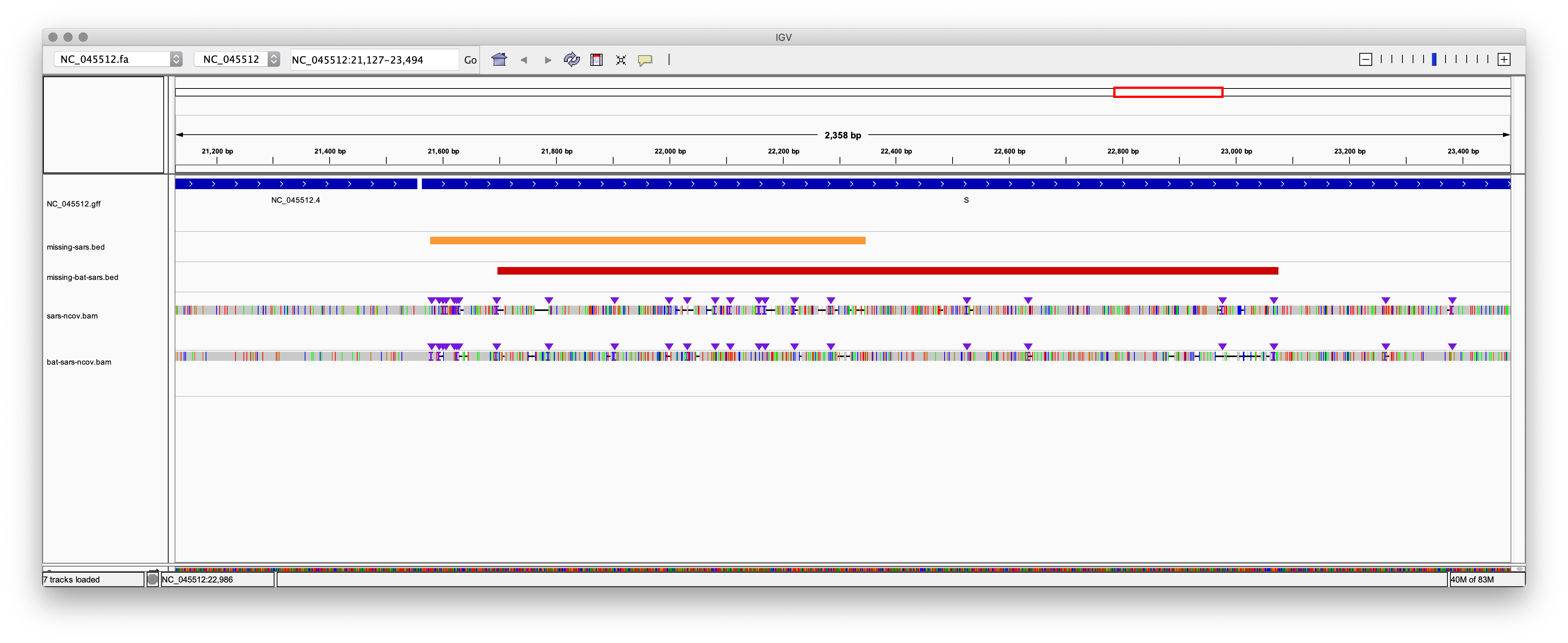
Caption: The primary region in the S surface glycoprotein is where
SARS, bat SARS, and the novel coronavirus diverge the most.
The goal of the book is to train readers in the arts of performing complex analyses, quickly, independently, and on their computers. We will demonstrate and explain what you can do, what type of results your analysis methods produce, and how you can interpret the information and draw informed and valid conclusions. The book is a unique take and perspective on the many challenges of genomic sciences while also providing hands-on solutions.
The new book and content is included with the Biostar Handbook.



@Alex Reynolds had provided this link in biostars slack: https://nextstrain.org/ncov , in case someone just wants to look at this type of data.
Hi Istvan, I have tried to download the ncov-sequences.yaml metadata as directed in your book but the ncbi page seems not to exist anymore. Could you help with how to go around this?
My apologies. Unfortunately, and without any notice, NCBI changed their page and even changed the format of the YAML that they now distribute from another location.
Doing so, they have made the previous, well researched and described process of obtaining the data invalid.
I am rewriting the book to be a little more generic to rely on genbank and taxonomy searches with solutions that would work for any other viral genome analysis in general (with SARS-COV-2 as a special case). It will take a bit of time, I will dedicate the upcoming week to that. In the meantime, I would recommend either downloading the last known data dump, or using the BLAST specific databases, those (for now) still work.
UCSC has a really well-constructed SARS-CoV-2 browser put together, which take a lot of data and makes it easy to explore and analyse against other datasets. There's a walkthrough of the browser and various tracks here: