Entering edit mode
7.3 years ago
Adele Feuerstein
▴
460
Why does the per base sequence quality decrease over the read in Illumina?
The first indicator for the quality of your sequencing data is the per base sequence quality of your raw reads. Often you will see a decreasing quality with increasing base position just as in the FASTQC image below. But what is the reason for this and what are the consequences?
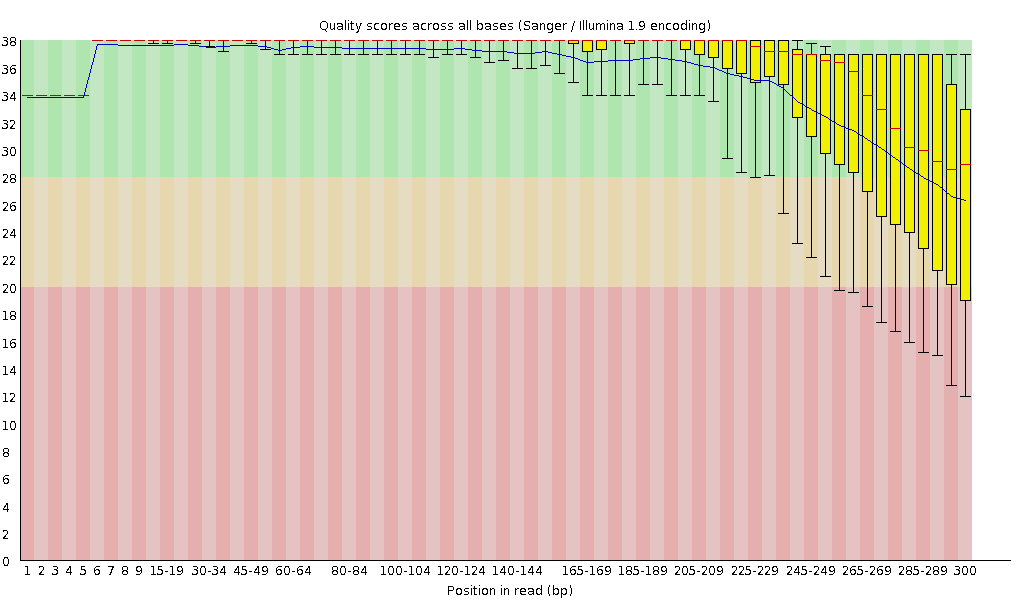


Even though reagents are held at 6-7C they still sit for the duration of the run. Cluster grow fatter over time leading to degradation in quality of basecalls. What you have posted looks like a typical MiSeq run.
As for consequences, it depends on what you are doing. As long as the median Q scores stay above 20-25 the sequence should be fine. If you are aligning to a known reference then you could afford to let Q scores go further down. If you are doing de novo work then you would want to start trimming at Q25 and below.
Did you look for the answer here or on other sites?, I can remember that this was asked very often already...
This post is a redirector for a blog entry at ecSeq (and not a tutorial hosted on Biostars).
I don't particularly like "stealth" blogs like this formulated as a question.
Interesting topic, though.