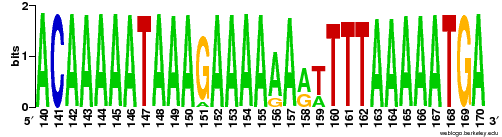
I have a SAM/BAM file and instead of displaying it in IGV or another browser, I would like to make a sequence logo out of it. My favourite software WebLogo does not support SAM/BAM file as an input (instead it supports CLUSTALW, FASTA, plain flatfile, MSF, NBRF, PIR, NEXUS and PHYLIP)
I would like to avoid parsing SAM file by myself and adding '-' when needed. Do you have any idea if there is already some software that could do such a thing? Thanks a lot.
The second question is about different alleles. I would like to visualize sequences (reads) from each allele with another color. I have found many coloring schemes, but none dividing sequences into groups together with corresponding coloring. Thanks a lot.


Pierre, you are definitely Tolkien of bioinformatics! Thanks.
Please help me in running sam4weblogo
I downloaded
jvarkit-master.zipand unzip how to use ityou need to compile it; Read the manual please.
I am having problems installing berkeleydb.jar, could you please give me short advice how to install it? (I am ubuntu user)
you don't need bdb, if you have filled the path to the picard jars. Just type:
should work.