Which program, tool, or strategy do you use to visualize genomic rearrangements?
In relation to my master thesis I'm working on tools to visualize fusion genes. In that regard I'm interested in any and all strategies and tools that exists for visualizing genomic rearrangements. I'll include the list I have so far below, and update the list when I get answers.
Programming packages:
- Circos: Perl package for circular plots, which are well suited for genomic rearrangements.
- J-Circos is a java application for doing interactive work with circos plots.
- ClicO FS: an interactive web-based service of Circos.
- rCircos: R package for circular plots.
- OmicCircos: R package for circular plots for omics data.
- ggbio: R package for visualizing biological data. Has a circular view similar to the previous packages.
- D3 chord diagrams (javascript) can be used to visualize genomic rearrangements. See this plot of migration flows as a similar example.
- chimeraviz: R package for chimeric RNA/fusion gene visualizations. (Disclaimer: I wrote this package.)
"Complete" tools:
- Genomatix Transcriptome Viewer: Gene Fusion analyses

- iFUSE: integrated fusion gene explorer

- FusionAnalyser: a new graphical, event-driven tool for fusion rearrangements discovery

- SOAPFuse includes the option to generate figures

- Gremlin

Gremlin is as far as I know not released anywhere. - Seqeyes: A flash tool for visualizing structural variations.


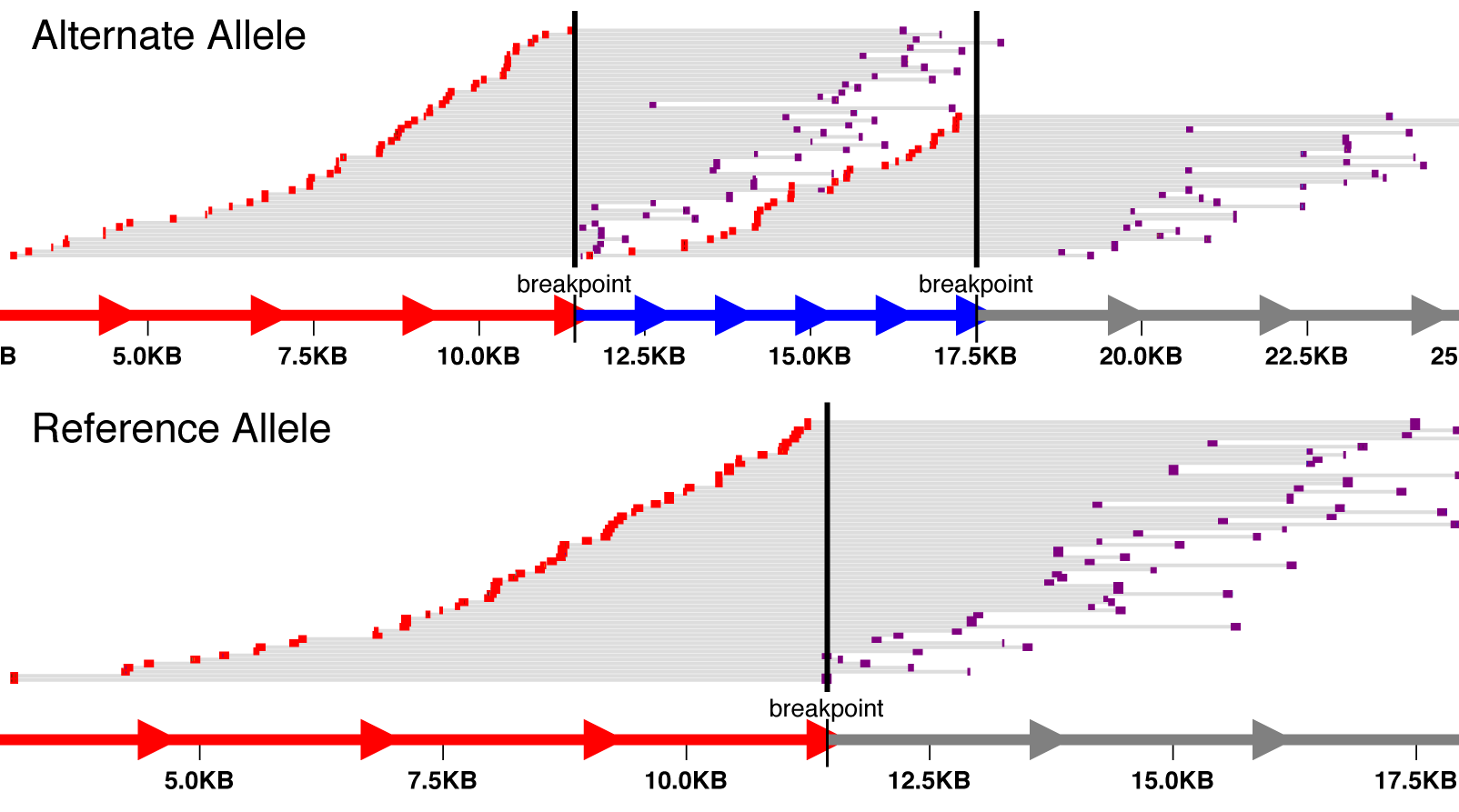

Just as a comment somewhat related to this: To the question A: I want to know what are some of the common problems facing a Bioinformatician? my answer is: Visualization!
Have you used, or know of, any of those tools to visualize circular RNAs? I am thinking shashimi plot for backspliced junction reads.