Some FASTA files (e.g., ESTs) have sequences with different IDs that nonetheless have the same sequence. I want to remove duplicate sequences based on the nucleotide sequence, rather than ID.HOW to do?Thankyou to all!
you can do this with fastx-toolkit
usage is like:
fastx_collapser < some.fasta > some.unique.fasta
linearize the sequences: surround the fasta headers by '@' and '#' , remove the CR, replace '#' by CR and '@' by '\t'
sort this tab delimited file on the second column (the sequence) , with case-insensible option, only the uniq columns
restore the fasta header and sequence
sed -e '/^>/s/$/@/' -e 's/^>/#/' file.fasta |\
tr -d '\n' | tr "#" "\n" | tr "@" "\t" |\
sort -u -t ' ' -f -k 2,2 |\
sed -e 's/^/>/' -e 's/\t/\n/'
I think this is the only answer that actually works if your fasta file contains a multiple sequence alignment.
Both fastx_collapser and gt sequniq fail because they consider '–' to be an invalid character in a fasta sequence. I didn't ask those tools to validate the sequences, and that's not their job, but they did it anyway making them useless for their actual purpose. Have these authors never heard of the Robustness Principle?
There are quite a lot of utilities that will do this; usually they are found as part of larger software packages (often aimed at motif discovery). For example, RSA-tools contains the utility purge-sequence, MEME contains purge. So those are some terms for your web search.
A roll-your own solution is quite easy if you store the sequences as hash keys (which have to be unique). For example using Bioperl SeqIO you could try something like:
use strict;
use Bio::SeqIO;
my %unique;
my $file = "myseqs.fa";
my $seqio = Bio::SeqIO->new(-file => $file, -format => "fasta");
my $outseq = Bio::SeqIO->new(-file => ">$file.uniq", -format => "fasta");
while(my $seqs = $seqio->next_seq) {
my $id = $seqs->display_id;
my $seq = $seqs->seq;
unless(exists($unique{$seq})) {
$outseq->write_seq($seqs);
$unique{$seq} +=1;
}
}
This will write out a new FASTA file, myseqs.fa.uniq, with only unique sequences (but no record of the other IDs with that sequence).
Depends on you application, but you may also try to filter out entries with the out same sequence but i.e. just 1bp shorter.
uclust --sort seqs.fasta --output seqs_sorted.fasta
uclust --input seqs_sorted.fasta --uc results.uc --id 1.00
uclust --uc2fasta results.uc --input seqs.fasta --output results.fasta --types S
This should give you results.fasta purged from identical but equal in size / shorter sequences.
Encode the sequences in hash using SHA-1 or MD5, and then check for collision. If you are familiar with Python, here is a useful recipe to start.
Here's a python script that should be able to do it:
from itertools import groupby
if __name__ == '__main__':
ishead = lambda x: x.startswith('>')
all_seqs = set()
with open(inname) as handle:
with open(oname, 'w') as outhandle:
head = None
for h, lines in groupby(handle, ishead):
if h:
head = lines.next()
else:
seq = ''.join(lines)
if seq not in all_seqs:
all_seqs.add(seq)
outhandle.write('%s\n%s\n' % (head, seq))
Wow, very clever. I don't fully understand a lot of the tool packages in python, but this is very intuitive. I modified the script slightly to strip the extra carriage returns in the outhandle.write command. This removes whitespaces that are aesthetically appealing but causes some programs to choke.
Works with big sets, aa/prot seq files (at the opposite to fastx-toolkit), very fast and simple to use: genometools:
gt sequniq -o out.fasta in.fasta
Already nice answers:
I would recommend to try the CD-HIT version designed for EST / nucleotides (CD-HIT-EST and CD-HIT-EST-2D) for this purpose.
uclust is about the fastest and one of the most flexible. If your sequences are not exactly the same length, or you also want to cluster some sequences that are almost, but not exactly, the same then look carefully at the flexibility and options of the thing you choose.
for smaller sets try this:
perl -ne 'BEGIN{$/=">";$"=";"}($d,$_)=/(.*?)\n(.+?)>?$/s;push @{$h{lc()}},$d if$_;END{for(keys%h){print">@{$h{$_}}$_"}}' multi.seq.fasta
and so readable too. ;) now which has "... all the visual appeal of oatmeal with fingernail clippings mixed in." ?? http://en.wikiquote.org/wiki/Larry_Wall
According to SeqAnswers: WU-blast, EBI-Exonerate, and bioperl all have stand-alone programs to make an "nrdb" = non-redundant database of sequences.
Note: WU-blast is now being distributed commercially as AB-Blast - get a free personal license. Older archived versions can be found here.
Strictly the nrdb program generates a non-identical database. The source for nrdb can be found in http://blast.advbiocomp.com/pub/nrdb/, no license required. This was a newer version than that bundled in WU-BLAST, no idea if this is still the case for AB-BLAST. Originally nrdb was used to produce NCBI's 'nr' and 'nt' databases. Of course 'nt' is no longer non-identical and 'nr' is produced using a slightly different process today, but nrdb is still a popular way to generate non-identical databases for use with BLAST.
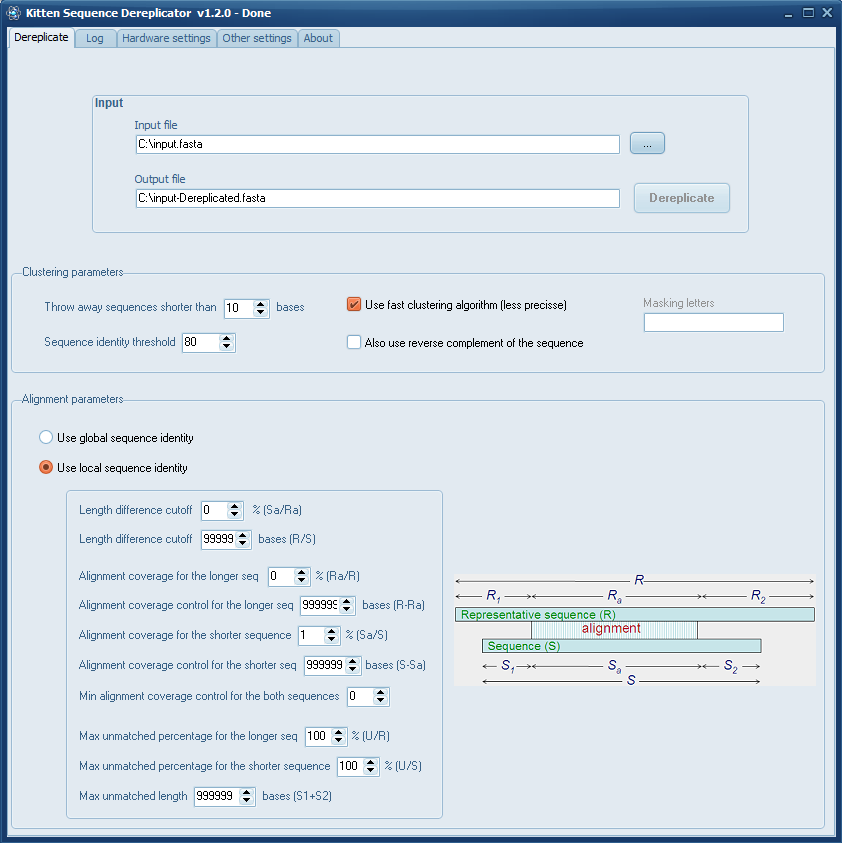
On Windows you can use my Kitten Sequence Dereplicator (which by the way, was updated recently).
The program is based on CD-Hit which is pretty accurate and fast.
I wrote a program for this problem, called Dedupe. It's very fast, and also (optionally) handles contained sequences. Usage:
dedupe.sh in=file.fasta out=nodupes.fasta
If you don't want fully-contained substrings removed, then add the flag ac=f (short for absorbcontainments=false).
I have to say that the remove of duplicate sequences is not at al trivial, especialy when you consider real data with sequencing/alignment errors. Also problematic is that sequences are identical, but do not sully overlap. I can say with certainty that CD-HIT-EST will filter some duplicates, but far from all.
I am working on a pipeline that uses BLAST, filtering out those contigs that have significant similarity to more that just themselves.
I tried fastx_collapser first, but it gives error for multiple aligned fasta sequences.
I found this useful website, which gives unique fasta sequences and concatenate the header name for the same sequences as well: http://www.ncbi.nlm.nih.gov/CBBresearch/Spouge/html_ncbi/html/fasta/uniqueseq.cgi
in R:
library(seqinr)
#when you dont use attributes to name sequences
fasnoa<-seqinr::read.fasta("fasta.fasta", set.attributes = F)
#get names
seqnames<-names(fasnoa)
#detect dups
dups<-grep(TRUE,duplicated(fasnoa))
#eliminate duplicates
namesnodups<-seqnames[-dups]
fasnoanodup<-fasnoa[-dups]
#write file
seqinr::write.fasta(fasnoanodup,namesnodups,"seqinrnoafasta.fas",nbchar=10000)
# when you use attributes as name of sequences
fas<-seqinr::read.fasta("fasta.fasta")
# read attribute annot when they have names and you will use them
namesfas<-lapply(fas, function(x) attr(x, "Annot") )
# delete attributes
for (i in 1:length(fas) ){
attributes(fas[[i]])<-NULL }
# detect duplicates
dups<-grep(TRUE,duplicated(fas))
# create object without duplicates
fasnodup<-fas[-dups]
# create object with names
namesnodups<-namesfas[-dups]
# modify names
namesnodups<-gsub("\\s+|,","_",sub(">","",namesnodups) )
# write file
seqinr::write.fasta(fasnodup,namesnodups,"seqinrfasta.fas",nbchar=10000)
seqmagick convert --deduplicate-sequences will remove duplicate sequence. See here:
http://fhcrc.github.io/seqmagick/convert_mogrify.html#examples
Hello Shaun:
I tried the seqmagic as you suggested, but it did not give what is expected.
>seq
ATCGATCGATATATATATAT
>seq2 part of seq1
CGATCGATATATATATA
>seq3 part of seq2
ATCGATATATAT
>seq4 reverse complementary of seq 2
TATATATATATCGATCG
>seq5 new seq
ATCGATCGACGATCGAGCGCG
>seq6 another new
ATCGATCGCGCGCGCGCGCGCGC
>seq7 psubstring of seq6
CGCGCGCGCGCGCGCG
Here is the command I used:
$ seqmagick convert --deduplicate-sequences test.fasta test_seqmagick.fasta
$ cat test_seqmagick.fasta
>seq1
ATCGATCGATATATATATAT
>seq2 part of seq1
CGATCGATATATATATA
>seq3 part of seq2
ATCGATATATAT
>seq4 reverse complementary of seq 2
TATATATATATCGATCG
>seq5 new seq
ATCGATCGACGATCGAGCGCG
>seq6 another new
ATCGATCGCGCGCGCGCGCGCGC
>seq7 psubstring of seq6
CGCGCGCGCGCGCGCG
Did I miss anything?
Here is my free program on Github: Sequence database curator
It is a very fast program and it can deal with:
- Nucleotide sequences
- Protein sequences
It can work under Operating systems:
- Windows
- Mac
- Linux
It also works for:
- Fasta format
- Fastq format
Best Regards
Try to use this python script, which can help you remove duplicates of the sequences in one or several fasta files.
And it's easy to use --id or --seq option to indicate whether you like the filter to work according to the id or the sequence itself.
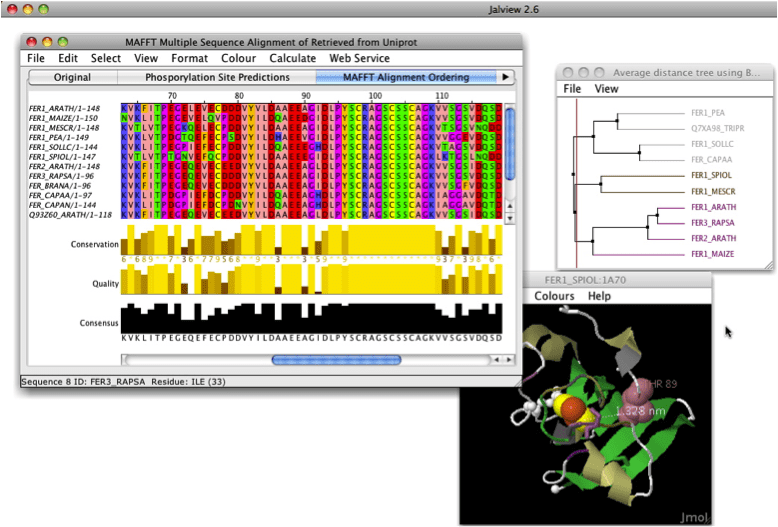
In Jalview (best alignment viewer) option "remove redundancy". You can select treshold.
Ctrl + D or edit > remove redundancy
 +
+
Use of this site constitutes acceptance of our User Agreement and Privacy Policy.


What is your understanding of "the same" are your including approximate stringmatching?
In fact you might not want to remove all the duplicated sequences but collapse them into a single sequence. I guess you meant to say it like this but your question is not clearly stating it.